
What is Restriction Endonucleases?
Restriction Endonucleases is known as restriction enzyme and also known as molecular scissors in biotech field. It cuts the DNA by identify 4-8 specific base pair.
They play a crucial role in the immune defense of these organisms against invading viruses known as bacteriophages.
Discovery
Initial Observation: Bacteriological researchers discovered in the late 1950s that some kinds of bacteria might fight off infection from bacteriophages, which are viruses that infect bacteria. They noticed that certain enzyme activity that may break the phage DNA were frequently acquired by bacteria that had survived a phage infection.
First Isolation: The Swiss scientist Werner Arber and his associates found in 1963 that the bacteria Escherichia coli (E. coli) contained an enzyme that could split foreign DNA into distinct pieces. This enzyme was given the term “restriction enzyme” because it prevented bacteriophages from growing by snipping their DNA.
Five years later, the first restriction endonuclease, called Hind II, was identified and characterized. Its activity was dependent upon a particular sequence of DNA nucleotides. It was discovered that by identifying a certain six-base pair sequence, Hind II successfully cut DNA strands at a precise location.
This specific base sequence is known as the recognition sequence.
As of right present, approximately 900 restriction enzymes have been identified from more than 230 bacterial strains, these enzymes are capable of identifying various recognition sequences.
Types
They are classified into several types based on their structural and functional characteristics. Here are the main types of restriction endonucleases:
Type I Restriction Endonucleases:

- Recognition Sequence: These enzymes recognize a specific DNA sequence, usually a palindrome, but the recognition sequence can be quite long (e.g., 4 to 8 base pairs).
- Cutting Mechanism: They cleave DNA at sites that are usually far (over 1000 base pairs) from the recognition sequence.
- Additional Subunits: Type I enzymes are complex enzymes with multiple subunits involved in DNA recognition, cleavage, and ATP hydrolysis.
- Example: EcoKI, EcoAI.
Type II Restriction Endonucleases:
- Recognition Sequence: These enzymes recognize short (typically 4 to 8 base pairs) palindromic sequences in DNA.
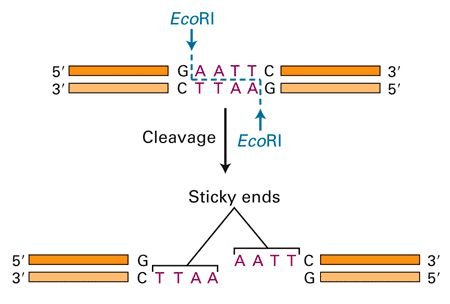
- Cutting Mechanism: They cleave DNA at or near the recognition site. The cleavage may result in blunt ends or sticky ends depending on the enzyme.
- Independence: Type II enzymes function independently of ATP and usually consist of a single protein unit.
- Example: EcoRI, HindIII, BamHI, etc. These are the most commonly used restriction enzymes in molecular biology research and biotechnology.
Type III Restriction Endonucleases:
- Recognition Sequence: They recognize specific DNA sequences like Type I enzymes.
- Cutting Mechanism: Type III enzymes cleave DNA at sites approximately 25 to 27 base pairs away from their recognition sequence.
- Coupling with Modification Enzyme: These enzymes work in conjunction with a DNA methyltransferase that methylates the same DNA sequence recognized by the restriction enzyme, protecting the host DNA from cleavage.
- Example: EcoP15I.
Mechanism of action
1.Recognition of DNA Sequence:
- Each restriction endonuclease recognizes a specific DNA sequence, known as the recognition sequence or restriction site. This sequence is usually palindromic, meaning it reads the same backward and forward (e.g., 5′-GAATTC-3′ for EcoRI).
- The enzyme scans the DNA molecule to find its specific recognition sequence, which typically consists of 4 to 8 base pairs.

2.Binding to DNA:
- Once the enzyme encounters its recognition sequence, it binds tightly and specifically to the DNA at or near this site. This binding is facilitated by interactions between amino acid residues in the enzyme and the bases in the DNA sequence.
3.Cleavage of DNA:
- After binding to the recognition sequence, the restriction endonuclease cuts the DNA backbone. There are two main types of cleavage patterns:
- Blunt Ends: Some enzymes cut both strands of the DNA molecule at precisely the same position, resulting in blunt ends with no overhangs.
- Sticky Ends: Other enzymes cut each DNA strand at a different point within or near the recognition sequence, creating single-stranded overhangs (sticky ends) that are complementary to each other.
4.Reaction Mechanism:
- Active Site: The enzyme has an active site where catalytic residues (usually amino acids) are located. These residues are responsible for the cleavage of the phosphodiester bonds in the DNA backbone.
- Mechanism of Cleavage: The enzyme typically uses two metal ions (such as Mg^2+) to coordinate the cleavage reaction, facilitating the nucleophilic attack on the phosphodiester bond.
- Water Molecule: A water molecule is often involved in the reaction, acting as the nucleophile that attacks the phosphate group of the DNA backbone, resulting in cleavage.
5.Release of DNA Fragments:
- After cleavage, the restriction endonuclease releases the DNA fragments. The size and nature of these fragments depend on the enzyme’s cutting pattern (blunt ends or sticky ends).
Applications
It have a wide range of applications in molecular biology, genetics, biotechnology, and various fields of scientific research. Here are some key applications:
1.Gene Cloning and Recombinant DNA Technology:
- Cloning: Restriction enzymes are essential for cloning genes and creating recombinant DNA molecules. They cut DNA at specific sites, allowing researchers to insert genes of interest into plasmids or other vectors.
- Vector Construction: Enzymes like EcoRI and BamHI are commonly used to generate compatible ends between DNA fragments and vectors, facilitating the insertion of foreign DNA into host cells for expression or replication.
2.DNA Fragment Analysis:

- Digestion Assays: Restriction enzymes are used to analyze DNA structure and function. By digesting DNA with specific enzymes, researchers can determine the presence and location of specific sequences within a genome.
- Southern Blotting: This technique involves digesting DNA with restriction enzymes, separating the fragments by gel electrophoresis, transferring them to a membrane, and detecting specific sequences using probes. It is used for DNA fingerprinting, gene mapping, and detecting genetic variations.
3.PCR (Polymerase Chain Reaction) Techniques:
- Site-Directed Mutagenesis: Restriction enzymes are used to introduce specific mutations into DNA sequences by cutting the DNA, followed by enzymatic repair with modified oligonucleotides during PCR.
- Restriction Enzyme Site Elimination: Enzymes can be used to remove or alter restriction enzyme recognition sites within a DNA sequence, facilitating subsequent cloning or analysis.
4.Genetic Engineering and Manipulation:
- Gene Editing: With the advent of CRISPR technology, which utilizes Cas proteins and guide RNA, restriction enzymes play a role in creating CRISPR constructs and targeting specific genomic loci for editing.
- Gene Expression Studies: Enzymes can be used to insert reporter genes into specific sites within genomes or expression vectors, allowing researchers to study gene regulation and protein function.
5.Molecular Diagnosis and Forensic Science:

- Diagnostic Tools: Restriction enzymes are used in diagnostic tests to detect genetic mutations or polymorphisms associated with diseases or genetic disorders.
- Forensic Analysis: DNA fingerprinting techniques, such as RFLP (Restriction Fragment Length Polymorphism) analysis, use restriction enzymes to generate DNA profiles for identifying individuals in forensic investigations.
6.Biotechnology and Industrial Applications:
- Production of Recombinant Proteins: Enzymes facilitate the production of proteins of interest by cloning genes into expression vectors and expressing them in host organisms.
- Bioremediation: Enzymes are used to manipulate microbial genomes for environmental cleanup by enhancing the breakdown of pollutants.
Challenges
They also present certain challenges and limitations that researchers need to consider:
Sequence Specificity:
- Limited Recognition Sites: Each restriction enzyme recognizes a specific DNA sequence, typically a palindromic sequence of 4 to 8 base pairs. This specificity can limit the choice of enzymes available for a particular application, especially if the desired DNA sequence lacks suitable restriction sites.
Cleavage Efficiency and Activity:
- Optimal Conditions: Enzyme activity often depends on specific buffer conditions (pH, salt concentration, temperature), and variations in these conditions can affect cleavage efficiency.
- Partial Digestion: Enzymes may sometimes produce incomplete or partial digestion of DNA, leading to heterogeneous DNA fragments that complicate downstream applications.
DNA Methylation Sensitivity:
- Recognition of Modified DNA: Some restriction enzymes are sensitive to DNA methylation patterns. Bacterial host DNA is often methylated to protect it from its own restriction enzymes. If the DNA to be manipulated is methylated differently, it may not be recognized and cleaved by the enzyme as expected.
Generation of Blunt vs. Sticky Ends:
- End Compatibility: Different enzymes generate DNA fragments with blunt ends or sticky ends (overhangs). Ensuring compatibility between the ends of DNA fragments is crucial for successful cloning and other DNA manipulations.
Limited DNA Fragment Size:
- Size Constraints: Some restriction enzymes have limitations on the size of DNA fragments they can cleave effectively. Very large DNA molecules may require special handling or alternative enzymes to achieve complete digestion.
Incompatibility with Some Sequences:
- Rare Recognition Sites: Enzymes that recognize rare or infrequent DNA sequences may be difficult to use due to the scarcity of suitable cutting sites within the DNA of interest.

FAQ (frequently asked questions)
1. What are restriction endonucleases?
- Restriction endonucleases, or restriction enzymes, are enzymes that recognize specific DNA sequences and cleave the DNA at or near these sequences. They are found naturally in bacteria and archaea as part of their defense mechanisms against viral infections.
2. How do restriction endonucleases work?
- These enzymes recognize specific DNA sequences (usually palindromic), bind to them, and cleave the DNA backbone. They can generate DNA fragments with blunt ends or sticky ends, depending on the enzyme.
3. What are the types of restriction endonucleases?
- Restriction endonucleases are classified into several types: Type I, Type II (most common), Type III, Type IV, and Type V. Each type has unique characteristics in terms of recognition sequence, cleavage pattern, and biological function.
4. What are restriction enzymes used for?
- Restriction enzymes are fundamental tools in molecular biology and biotechnology. They are used for DNA cloning, creating recombinant DNA molecules, analyzing DNA structure, gene editing, and various other applications in genetic research and biotechnology.
5. How do I choose the right restriction enzyme for my experiment?
- Choose an enzyme based on the specific DNA sequence you need to cleave. Check the enzyme’s recognition sequence and cutting pattern (blunt ends vs. sticky ends). Consider factors like buffer compatibility, enzyme activity at specific conditions, and any potential sensitivity to DNA methylation.
6. What are the challenges of using restriction enzymes?
- Challenges include limited availability of suitable recognition sites, enzyme specificity and activity under optimal conditions, sensitivity to DNA methylation, potential for partial digestion, and limitations on DNA fragment size.
7. What is the difference between blunt ends and sticky ends?
- Blunt ends are generated when a restriction enzyme cuts DNA straight through both strands at the same position, leaving no overhangs. Sticky ends are produced when the enzyme cuts each DNA strand at different points near the recognition sequence, resulting in single-stranded overhangs that can base-pair with complementary ends.
8. Are there alternatives to restriction enzymes?
- Yes, alternatives include CRISPR-Cas systems for genome editing, zinc finger nucleases (ZFNs), and transcription activator-like effector nucleases (TALENs). These technologies offer more precise control over DNA manipulation and editing compared to traditional restriction enzymes.
Conclusion
In conclusion, restriction endonucleases stand as pillars of genetic engineering, enabling precise manipulation of DNA with profound implications for science and society. From their humble origins in bacterial defense mechanisms to their current role as indispensable tools in laboratories worldwide, these molecular scissors continue to shape the forefront of biotechnology.
Table of Contents
Read more about Restriction Endonucleases
Bt cotton – types, advantages, challenges
Go and visit dusearchit.in and get more knowledge about others topics.
